A Review of the Central Dogma
17 Nov 2015This post is here to provide a basic overview of the central dogma of molecular biology as a introduction for a later post on Haskell functions pertinent to the mechanisms in the central dogma. To get to that post, click here.
If you’re interested in a general review of the central dogma, its mechanisms and current place in modern biology, continue reading. Feel free to skip it if all of that is old news.
This review assumes some basic familiarity with the base-pairing nature of nucleic acids and the structural nature of DNA, RNA, and protein.
What the “central dogma” is all about
The central dogma of molecular biology is the codification of how information is transferred between the biopolymers: DNA, RNA, and protein. It is sometimes given in the simplest form as “DNA proceeds to RNA proceeds to protein”. To put it in other terms, the central dogma as a statement expresses the idea that base-sequence information is conveyed from DNA, to RNA, ultimately to protein, and sequence information in protein is never conveyed back to DNA or RNA. Here’s a little diagram of what the central dogma allows:
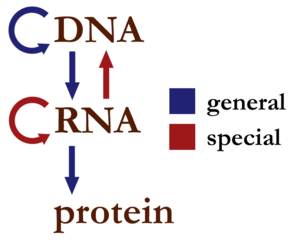
Much has been discovered since the central dogma was originally formulated, giving us a richer view of genetics and biology. In light of some of these discoveries, some argue that the central dogma has been falsified, leaving it perhaps only useful as a general statement of how most things work. I believe that these views frequently reflect how our thinking has shifted with regards to information in a biological context. There’s a lot more to information than solely the sequence information with which the central dogma was originally concerned.
Falsification of the central dogma?
What are the falsification conditions of the central dogma? If any of the following were demonstrated, we can say that the central dogma has been demonstrated false:
- Protein to Protein information flow
- Protein to DNA information flow
- Protein to RNA information flow
If the decoding of protein sequence to nucleic acids (reverse translation) were to be discovered, that would be a refutation of the central dogma. So far, no such discovery has been made. However there is some debate over whether prion activity represents a violation. This paper and the conversation that follows provides a good discussion of the matter. I’ll try to give a short layperson’s summary.
A prion is a protein which may switch to a different folded shape and further induce others to do the same. This is the basis of some neurodegenerative diseases as well as a form of epigenetic heritability in some organisms. One particularly intriguing case is that of prion activity affecting how the translation of many other proteins takes place, the ribosome does not terminate as typical, and so affects the sequence information of other proteins. In light of this, even keeping to a very strict sense of the central dogma, it could be argued that protein sequence information may result in altering protein sequence information and violate the dogma.
I’m not sure whether or not I am fully decided here, because it would seem to me that there is little distinction between this subject (though certainly no less fascinating) and other epigenetic factors. I might suggest then that if epigenetics do impinge on the central dogma, then it is contradicted not only by prions, but other epigenetic modes as well that influence biopolymer sequence.
Whether or not the central dogma stands does not undermine the foundations of biology. If falsified, the central dogma may remain useful as a negation which manages to capture a great quantity of known molecular biology. Either way, it is vital to note that one’s understanding of biology does not end with the central dogma.
Mechanisms of sequence information flow
Now I’ll turn the attention over to known mechanisms of information flow in the central dogma with some examples of sequence transformations.
DNA : {A,C,G,T}
DNA to DNA occurs ubiquitously, replication of DNA by DNA
polymerases is essential for providing genetic material to an organism’s’
progeny. Fidelity of replication is important so that viable organisms produce
enough viable progeny, as is the occasional infidelity to provide diverse
progeny that might respond better to change and competition. Replication of
GATTACA would give GATTACA, with possible chance mutations.
DNA to RNA occurs ubiquitously to produce RNAs of a very wide wide variety
of functions. The process of producing RNA from DNA templates is known as
transcription and is carried out by RNA polymerases that create RNA chains in
a pair-wise complementary fashion to the DNA template. RNA is chemically very
similar to DNA, one distinguishing feature is the use of the nucleobase uracil (U)
in place of the nucleobase thymine (T) of DNA. Both are complementary to the
nucleobase adenosine (A). Thus the corresponding RNA sequence of GATTACA,
perhaps transcribed from the complementing strand would be GAUUACA.
DNA to Protein is not known to occur in nature, though it has been
accomplished in vitro under certain conditions
It is not forbidden by the central dogma, and there is speculation as to why it
is either rare or non-existant. It is important to address that we frequently
refer to the genetic code, the
coding of amino acid sequence for protein by nucleotide triplets, in DNA terms
even though it must first pass through a transcribed mRNA intermediate (more on
that below). I’ll extend GATTACA up to 9 nucleotides for this example:
GATTACAGA would translate to the amino acid sequence DYR. Be aware that the
genetic code can vary in some cases.
RNA : {A,C,G,U}
RNA to DNA occurs in many organisms and was considered a possibility from
the beginning of the central dogma, though evidence for it came somewhat later.
RNA is used as the basic genetic material for some viruses; retroviruses, HIV is
one example, must reverse transcribe their RNA genetic material to DNA before
it is integrated into the genome of the host cell. RNA templates are used in
reverse transcription to extende the telomere caps of chromosomes by
telomerases. To do reverse transcription, just reverse transcription! GAUUACA
produces GATTACA.
RNA to RNA replication occurs via RNA-dependent RNA polymerases sometimes
called replicases. This is important to the replication of viruses with RNA as
genetic material as well as eukaryotic cells for RNA silencing.
RNA Silencing is a huge deal in biology, not just for its natural role, but
because it serves as a powerful tool of basic research. Ignoring stochastic
mutations, base complementarity means that GAUUACA will give GAUUACA.
RNA to Protein is conducted by a massive complex of protein and RNA called
the ribosome, using the genetic code
that was mentioned earlier, as it scans over a messenger (mRNA) template. This
is the only known way to produce protein and is called translation.
Translation from the RNA sequence GAUUACAGA in the (not totally) universal
genetic code would produce the amino acid sequence DYR.
Protein : {A,R,D,C,Q,E,G,H,I,L,K,M,F,P,S,T,W,Y,V,...}
Protein to DNA or RNA is not known to occur in nature, in fact it is
strongly suspected to not occur. This is not permitted to occur according to
the central dogma. There’d be a Nobel Prize for whoever discovered a natural
biological system that accomplished this. It might then be interesting that this
is a common research job. Before genome sequencing was developed and became
relatively cheap, protein sequencing
might have yielded the amino acid sequence of the protein, which would need to
be reverse translated through the genetic code to either get an idea of what
the encoding gene looks like (introns and other gene features are lost data), or
to produce the protein transgenically. Protein sequencing is still an important
part of biological research, predictions made in bioinformatics on genomic
sequences need experimental validation after all. Reverse translation is made
somewhat complicated by the fact that the genetic code is degenerate, meaning
that amino acids may be coded by more than one codon, so there are many
nucleic acid sequences that could hypothetically produce the same protein.
To be brief, the choice of codon can affect how well a sequence is translated
or what other factors may regulate the mRNA; this is an area of active study.
One rudimentary approach to codon optimization is
to calculate the relative frequency of codon usage in an organism’s
protein-encoding genes and choose common codons rather than underrepresented
ones. For the sequence DYR in the universal genetic code, we would have the
following (brackets indicate choose one) [GAT,GAC][TAT,TAC][CGA,CGC,CGG,CGT,AGA,AGG]
for a total of 2 * 2 * 6 = 24 possible corresponding DNA sequences.
Protein to Protein As mentioned earlier, prions may represent a form of protein sequence modification by other proteins and it is a matter of current some discussion whether or not this is a refutation of the central dogma that excludes protein to protein sequence transferrence. Protein is not replicated from protein, nor is it produced by other means aside from translation.
On a final note
I did not mention that DNA, RNA, and protein can be modified in ways beyond their sequence, but it is very commonly so. In addition they interact with eachother and with various components in their biological system. Much of this is outside the concern of the central dogma, but entirely within our concern as scientists seeking to understand the world. So the central dogma is really a Bio 101 concept, from there we have moved on.
The wonderful and bewildering nature of biology is that it is complex. It is the interplay of numerous agents that gives rise to the diversity of the world in which we live, why biological research remains difficult, why some diseases have no simple cures.
For some further reading on the complexifying factors of general information flow in biology, the following key terms come to mind and in no particular order: epigenetics, intron excision, alternative splicing, RNA editing, RNA silencing, post-translational modification, inteins, riboswitches, horizontal gene transfer.